This is the website for the code and user documentation of the FindSim project. The published paper is here. The related FindSimWeb project provides a web interface to search, view, edit, and run FindSim experiments.
FindSim is a Python/MOOSE framework to map experimental protocols and readouts to models of neurophysiology and cellular signaling. FindSim simulates the experiment, checks how it fits the data, and provides tools for parameter analysis and optimization. These are designed to run from the command line, and in batch mode for large parallel optimization runs.
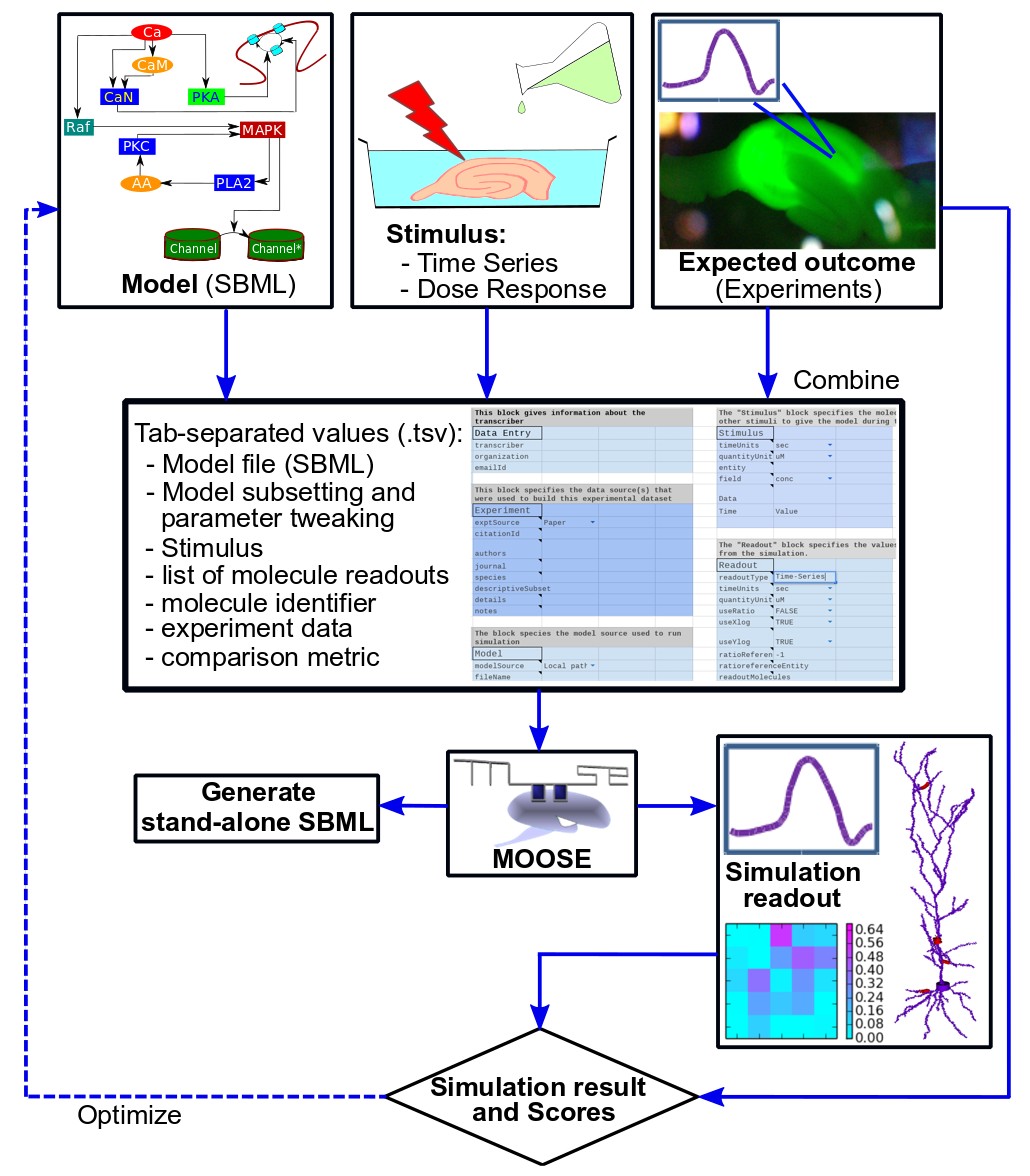
The workflow for FindSim is to identify/build a model, select an experiment, codify it, and run the experiment on the model. With the additional tools one can do a parameter exploration and sensitivity analysis, and single or multi-parameter optimization.
